TrajectoryNet: A Dynamic Optimal Transport Network for Modeling Cellular Dynamics
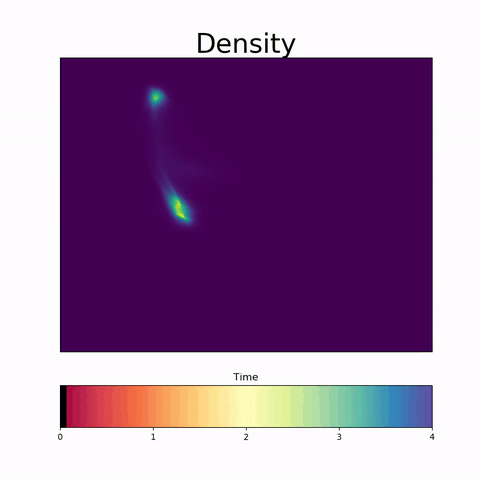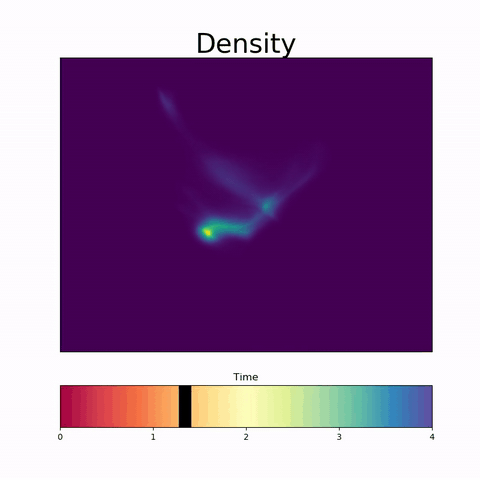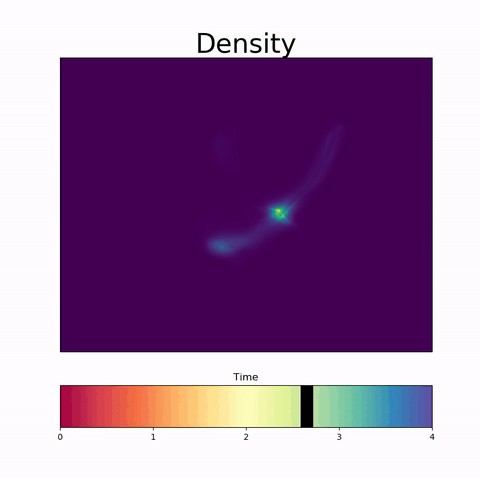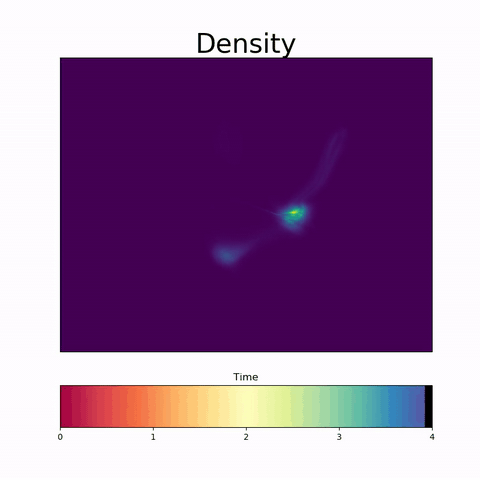 Modeling Embryoid Bodies over time from cross-sectional single-cell RNA sequencing.
Modeling Embryoid Bodies over time from cross-sectional single-cell RNA sequencing.Abstract
It is increasingly common to encounter data from dynamic processes captured by static crosssectional measurements over time, particularly in biomedical settings. Recent attempts to model individual trajectories from this data use optimal transport to create pairwise matchings between time points. However, these methods cannot model continuous dynamics and non-linear paths that entities can take in these systems. To address this issue, we establish a link between continuous normalizing flows and dynamic optimal transport, that allows us to model the expected paths of points over time. Continuous normalizing flows are generally under constrained, as they are allowed to take an arbitrary path from the source to the target distribution. We present TrajectoryNet, which controls the continuous paths taken between distributions. We show how this is particularly applicable for studying cellular dynamics in data from single-cell RNA sequencing (scRNA-seq) technologies, and that TrajectoryNet improves upon recently proposed static optimal transport-based models that can be used for interpolating cellular distributions.
Also at LMRL Workshop @ NeurIPS 2019